tensorICA (Tensorial Independent Component Analysis)
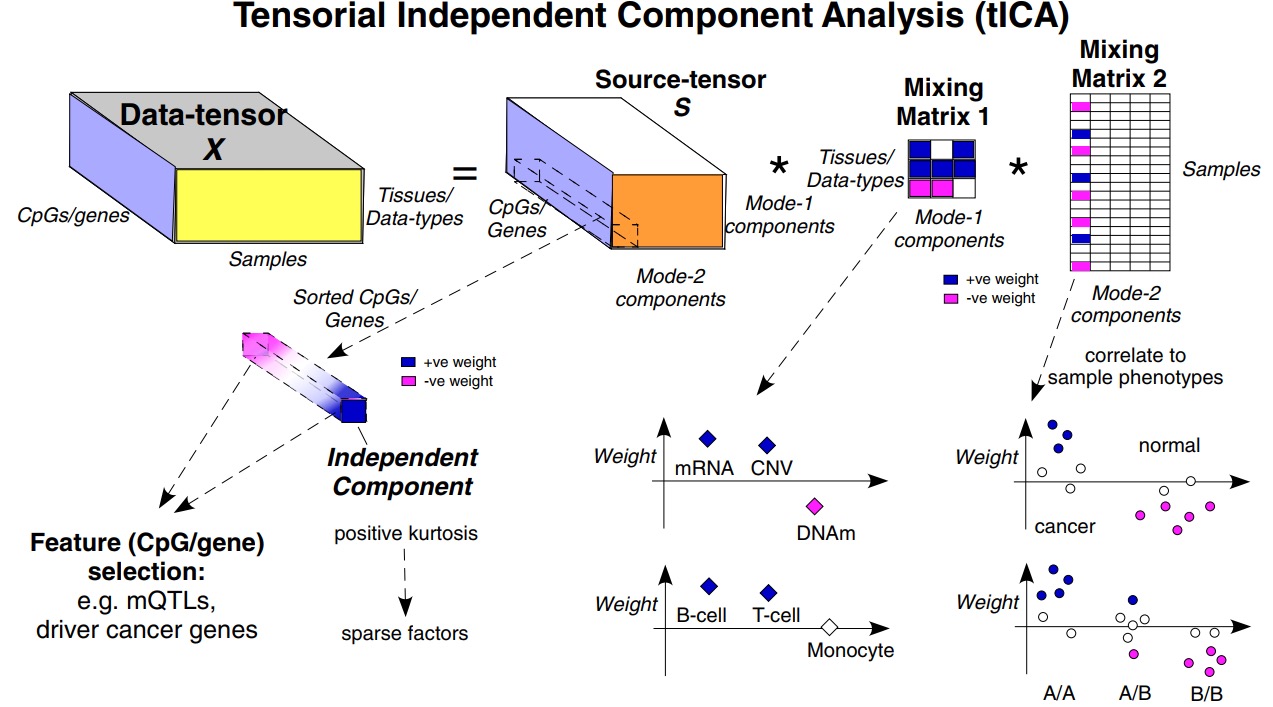
TensorICA (Tensorial Independent Component Analysis) package is designed for fast decomposition analysis of tensor-valued multi-omic data. It contains several functions for decomposing multi-omic tensor-valued data implementing two tensorial ICA methods and several utility functions for visualizing the relationship between component against phenotype to help with feature selection. Tensorial ICA aims to infer from a data-tensor, statistically independent sources of data variation, which should better correspond to underlying biological factors. Indeed, since biological sources of data variation are generally non-Gaussian and often sparse, the statistical independence assumption implicit in the ICA formalism can help improve the deconvolution of complex mixtures and thus better identify the true sources of data variation. In the package, we here include two tensorial methods call JADE and FOBI.
Maintainer: Han Jing
Citation
Teschendorff AE, Han J, Paul D, Virta J, Nordhausen K. Tensorial Blind Source Separation for Improved Analysis of Multi-Omic Data. Genome Biology (2018) 19:76.
Installation
To install this package, start R (version "3.6") and enter:
if (!requireNamespace("devtools", quietly = TRUE))
install.packages("devtools")
devtools::install_github("jinghan1018/tensorICA")
Documentation
PDF: tensorICA Vignette